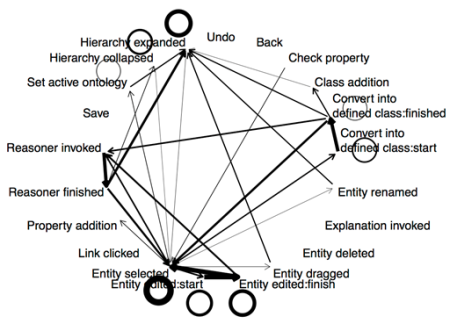
We have finally published our paper on the Minimal Information for Reporting an Ontology (MIRO). We have spent years on this paper; events have intervened to cause the marathon. The paper’s details are
MIRO: guidelines for minimum information for the reporting of an ontology
N Matentzoglu, J Malone, C Mungall, R Stevens
Journal of biomedical semantics 9 (1), 6; 2018.
The motivation for this came from the observation that the many ontology description papers published did not make a consistent, good description of the ontology in question. It is easy to make the observation, but less easy to say what should be contained in a “here’s my ontology” paper.
This is not a matter of reproducibility – one shouldn’t expect to take a method from an ontology paper, follow that method, and get the same ontology. Perhaps roughly the same ontology – same topic area, same naming conventions, same patterns, etc. etc., but the exact same ontology isn’t possible. One should be able, however, to read an ontology description paper and understand the topic, scope, content and development process for an ontology. There isn’t much substitute for looking at the ontology in some form, but the report should be able to convey most of what one needs.
The ultimate goal is community involvement in shaping and then following the MIRO guidelines so that ontology reports are adequate, so the ontology community should be involved in what the MIRO contains. This is an outline of what we did:
- James Malone, Chris Mungall and I drafted the guidelines.
- We put them up in an on-line survey tool.
- We gathered input from the community via this survey
- WE updated the MIRO guidelines
- We then selected some extant ontology reporting papers and analysed them to see how well they complied with the MIRO guidelines.
The MIRO guidelines can be seen on their GitHub repository and the on-line survey still exists. The main sections of the MIRO guidelines are:
- Basics – name and so on.
- Motivation – why this ontology?
- Scope, requirements etc. – what should be in the ontology.
- Knowledge acquisition. – How was the ontology content obtained?
- Ontology content. – What’s in it and how is it represented?
- Managing change – obvious.
- Quality assurance. – is it any good?
At the time of the survey’s analysis we had had 110 responses that, given the size of the bio-ontology community, is a pretty good response. (MIRO is not focused on the bio-health community, but that community is such a big player and I’m part of it, so it’s a good yardstick.)
Here is not the place for the details of the survey response, it can be seen in the paper itself and via the RPubs site for MIRO. Overall, however, we had a very positive response to our proposal.
In the survey, we only gave the names of the reporting guideline; we did not operationalise the guideline. This was deliberate: First to get focus on the guideline, its name etc. and second so that comments could be used to add to the existing operationalisations without too much bias as to how that operationalisation should be done. We got much in the way of useful input on names and on how to operationalise the guidelines. There were, however, no substantial changes to the MIRO guidelines themselves.
One bit of community feedback is that everything (almost) in the MIRO guidelines are mandatory and this may be too onerous. I make no apologies for this – MIRO is minimal; thus it has only or nearly only that which is essential – most optional things should be beyond the minimal. Nevertheless, complying with the MIRO may be an onerous task, but sensible development techniques may make the task less onerous.
Two items that were deemed to be of less importance surprised us:
- Ontology metrics – numbers of classes, axioms and so on. This is easy to compute and gives some idea of scale – though obviously no detail – of an ontology. It may be that readers of ontology reports find these kinds of numbers less useful or easy enough to find by looking at the ontology.
- How the content was chosen- this is not just scope, but also priority (though we asked for that exercise to be reported too). An explanation of what is in and out of scope would seem important – it may be, as we say in the paper, that scope is implied by the requirements and so on.
Our review of ontology papers showed that only 41% of the MIRO items were covered in the ontology papers. Some of the main areas in which ontology reporting papers didn’t comply with MIRO were:
- Testing the ontology – not an easy task, but even simply reporting that the ontology has no unsatisfiable classes and can answer queries that show its core competence would be a good start.
- Evaluation reporting was scant – is it the ontology that people want in the way that they want it?
- Versioning.
- Sustainability – will the ontology carry on being developed?
- Entity deprecation policy – as the ontology develops and entities change, is there a process involved?
- Metadata policy – what metadata, usually decorating a class – is supplied; this was not well reported.
- The ontology’s licence; surprisingly poorly reported. Here we did not check whether the ontology was released with a licence.
The survey of compliance generally supports the original motivation for the MIRO guidelines – that ontologies are not well reported in the literature. We’ve also had good input into shaping the MIRO guidelines; all we need now is adoption…. The MIRO guidelines should be adopted and most of the information features are generally easy to report. Support for MIRO by the community should make ontology reports better and I recommend them to the community.