Our BMC Bioinformatics paper on Populous has been published. This first had an outing in the 2010 SWAT4LS workshop. Populous is Simon Jupp’s spreadsheet based tool for “populating” or adding content to ontologies. It builds upon the use of spreadsheets as templates for filling out what we know about entities and then transforming that information into axioms within the ontology – we just think that Populous does it rather well. The thing about Populous is that it isn’t just filling in bits of OWL in a spreadsheet; it is set up to hide as much of that as possible, as a way of getting non-ontologists close to ontology building without “frightening the horses”.
We used Populous in the construction of the KUPKB. We needed to extend the Kidney and Urinary Pathway Ontology (KUPO) to include more kidney cells that were not in the Cell Type Ontology, but Julie Klein, one of our collaborating kidney specialists was not and would not be an OWL or Protégé user – even if we supplied the template of axioms for her to fill in. This is why we went for spreadsheets as a way of collecting the classes that go into the predefined framework of properties. For the KUPKB, we wanted to describe the anatomical location of the cells and the biological processes that they fostered. This is a simple template to put in a spreadsheet, but leaves the problem of which classes to put into the spreadsheet’s cells – we don’t want users having to go back and forth to the GO (or whatever) to choose cells, transcribe or cut-and-paste the ID or just make up terms as all this is too much like hard work.
This is where Populous comes in; it supports the scenario above, but puts menus in place that have the appropriate terms from the “supporting” ontologies in place. Populous also has support for OPPL, the Ontology Pre-processing Language, that takes rows from the spreadsheet, creates the axioms and squirts them into the growing ontology via the OWL API.
how populous works
Populous is an extension of RightField. This is a Manchester product from Carole Goble’s group based on spreadsheets. RightField is a spreadsheet generator. RightField is used to choose portions of vocabulary to go into a column; these are presented as menus to a user in the Excel spreadsheet that RightField produces. RightField manipulates Excel’s ability to create such menus. The generated spreadsheets are then handed out to biologists to use. The important point is that it is just using an Excel spreadsheet – no other software need be installed (a barrier to use and, of course, another maintenance issue). RightField was developed for systems biologists to standardise their annotations of data. It is “semantics by stealth” as the whole idea is to make it easier to do the right thing (use terms from a vocabulary), rather than just making terms up (because that is easier than looking for the terms in a vocabulary). In RightField, the spreadsheet designer has already chosen the right area of vocabulary to be used. It doesn’t stop wrong or new terms being used (after all, vocabularies are rarely complete). However, it does afford a mechanism for spotting non- vocabulary terms and dealing with them – either changing the spreadsheet or fixing the ontology itself. RightField help data to be marked up; it can then be transformed to whatever one wants.

Figure 1. A RightField spreadsheet showing one of the “ontology driven” menus
The picture in Figure 1 is of RightField showing one of the "ontology menus" for describing some data that went into the http://www.kupkb.org[KUPKB]. In the first column, we describe what type of metadata we want; e.g. compound list, experiment condition, species In the second column, the cells are yellow, they are the actual RightField constraint cells. For example, for the metadata about maturity we can see that the cell expands in a menu where the user can choose amongst the appropriate ontological terms: adolescent, adult, fetal
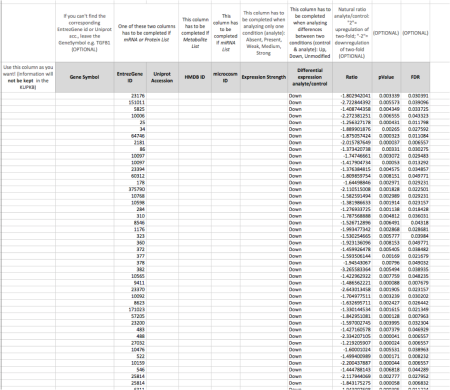
Figure 2. A RightField spreadsheet showing some annotated data
The picture in Figure 2. shows some data inside the RightField spreadsheet; it is some experimental KUP data that went into the KUPKB. We can just copy/paste lots of tabulated data into the spreadsheet cells. Here, for example, we have a list of genes, identified with their EntresGeneId (so the list of identifiers has been pasted into the EntrezGeneId column). But if you have other identifiers, such as GeneSymbol, UniprotId, HMDBId, or microRNAId, you can just copy/paste in the appropriate column. Then you have a column to describe how each gene is modified in the disease; this is the Differential expression column and for each gene you can say if it is Up or Down regulated – offered via ontology terms from an Excel menu. Finally, there are 3 columns to enter numerical values such as Ratio, pValue and FDR (False discovery Rate).
Populous re-purposes RightField to make it do ontologies. The mechanism is basically the same, but Populous adds a pane for the OPPL scripts to be made to transform the spreadsheet’s contents to OWL axioms and put them in the target ontology (there is a wizard to take one through the process of mapping columns to variables and so on). Instead of annotating data, we’re annotating the entities to appear in an ontology. For the Kidney and Urinary Pathway Ontology (KUPO) we made a Populous sheet for cells (we needed to augment the Cell type Ontology with Kidney cells). We wanted a patterns something like:
Class: KUPOKidneyCell
SubClassOf: Cell,
isCapableOf some biologicalProcess,
isPartOf some GrossAnatomicalEntity
|
(though that isn’t exactly what we ended up with.) So, the left-hand column of the sheet is the biological cell; the next two columns are the isPartOf and the isCapableOf property and the entries in these columns are the existential fillers for these properties.

The KUPO Excel spreadsheets can be taken away and filled in (for KUPO Julie took them away and added information about kidney cells). The picture above shows some kidney cells being ontologically described in Populous. Each column is filled with terms from one of the ontologies used for KUPO. In the first column, this is terms about Cell Types using the Cell Type Ontology. Most of the terms are red because we had to create them; they were not present before in CTO. Only podocyte and juxtaglomerular cell are in green because they existed in CTO. In the third column it is terms about Anatomy’ using the Mouse Anatomy Ontology. Here it is the opposite, most of the terms are green (i.e. existed in MAO) except for renal corpuscule and bowmans capsule. In the last column are terms about Biological Processes using GO – and there all are validated “green”. An export of a populated KUPO spreadsheet can be viewed here.
On loading into Populous, items in spreadsheet cells not in the chosen vocabulary are highlighted in red and OPPL has a way of dealing with such “new terms” – after all, we do want to collect new vocabulary, though this highlighting does allow some errors to be fixed. The code below shows the OPPL pattern that actually takes items from the spreadsheet, puts them into the variables, fills in the pattern template and puts them, via the OPPL and OWL API into the ontology (in this case KUPO).
?cell:CLASS, ?anatomyPart:CLASS, ?partOfRestriction:CLASS = part_of some ?anatomyPart, ?anatomyIntersection:CLASS = createIntersection(?partOfRestriction.VALUES) BEGIN ADD ?cell equivalentTo CL_0000000 and ?anatomyIntersection END; |
The OPPL script (at its most basic) adds words like add, remove and select to the Manchester OWL Syntax; it then has its own interpreter and runs the OPPL script, via the OWL API, on a given ontology. In Populous, we have a series of variables mapped to columns; each row is taken in turn and added to the script, which then adds the appropriate axioms to the ontology.
(Populous also does lots of sensible things about identifiers, labels, multiple values in cells and so on.) A slightly out-of-date Populous video can be viewed here.
why spreadsheets are good.
By using Populous we had Julie Klein, our collaborating biologist, ontologically describing cells just by choosing items from a menu to fill a pattern implicit within the spreadsheet she was using. Julie added some 183 kidney cell descriptions to KUPO without pressing a single button in Protégé .
Why is this technique a good thing? first of all it uses spreadsheets. the Excel spreadsheet is almost ubiquitous in science; so just join in. This is what RightField has done – trying to get one’s user base to install another bit of software, and one with which they are unfamiliar, is putting oneself onto the backfoot from the start. by using the Excel spreadsheet, we’re adopting a familiar environment. we just dish out the spreadsheet and that’s it. the Populous and OPPL bits can be done off-line and by someone else.
We can also change the patterns by which we add the descriptions to an ontology by changing the OPPL script. OPPL is a means by which the ontology can be transformed programmatically without resorting to the depths of the OWL API – and this is also a good thing. We can also automatically add all that tedious but important stuff like author tags (though I suspect we didn’t), but we should have done.
Third, we have a record of what was added; we can expand the spreadsheet with relative ease and just add more stuff to the ontology.
Most of the KUPO was built with few button presses in Protégé . Simon Jupp built the framework from OBO ontologies, plus a few bits of our own. KUPO extensions and the experimental data that form the knowledgebase are added using RightField and Populous. There were few button presses and this is a good thing. It reduces the possibility for error and it makes the use of patterns ruthlessly consistent. However, the principle thing is that we had Julie really contributing to the ontology, but she didn’t have to do OWL, Protégé or “ontologising”.
One thing missing from the paper is an evaluation. The problem is we didn’t really know what to evaluate. We could have sat down some biologists (that didn’t know how to use OWL or Protégé ) in front of an Excel spreadsheet and Protégé , ask them to describe cells and measured the result. I suspect Julie’s reaction (and others) would have been “bof”. This would have been a pointless thing to do. We could have tested the usability of Populous/RightField, but that’s not the claim we’re making – we’re talking about making building ontologies “easier” – always a bad word to use, but it’s what we’re doing. We know that it works from a technical point of view, but that’s a bit ordinary. We could compare manual building against auto- building to look at error or slip rates. this is hard to measure (except with Eleni Mikroyannidi’s regularity Inspector for Ontologies); this would be useful, but again isn’t really testing the claim that it is easier for non-ontology builders to use something like a spreadsheet.
We’re continuing to use Populous. we recently ran an event expanding the Software Ontology; Duncan Hull made a RightField spreadsheet with which attenders could use the SWO to describe software, then he wrote OPPL to add the stuff to the SWO. Our attenders were not ontologists and so, again, we had domain experts directly contributing to axiomatic descriptions of classes in an ontology without the trauma of “doing ontology” – which must be a good thing.
(this blog had lots of input from Simon Jupp and Julie Klein)