Protege is a complex tool. By deafult it offers a user all that it is possible to do with OWL (relationships, quantifiers, class expressions, and so on) and there can be choices at any point. Often Protege is too complex, especially at an early stage of authoring an ontology where one might simply wish to “sketch” something out; perhaps for migrating to a more sophisticated form later. Andrew gibson, when he worked in our group, wanted a simple tool for “sketching” an ontology. Such a tool would be based on a simple “blob and line” model, corresponding to classes, subclass axioms and existential restrictions. Matt Horridge developed a tool called Montage from Andrew’s specification; it was only ever a prototype and never saw the light of day outside that particular office.
Inspired by this, I offered a third year undergraduate project to develop a “simple knowledge organisation tool” — SKOT. A student called Mark Jordan took this project on and has done a good job, given the restrictions of time involved in a University of Manchester Computer Science third year undergraduate project.
Mark developed a tool called SKOT – the Simple Knowledge Organisation Tool. It is an open source project under an LGPL licence and the SKOT code is available on source forge.

The picture shows SKOT at a point just before the user is about to export the sketch into an OWL file. There is a “term list”, where words or terms that might form the blobs or classes in the ontology are “stored”. The example is the traditional “university” modelling example, with the terms “University”, “Person”, “Student”, “Undergraduate”, “Mark”, “Postgraduate”, “Teaching Assistant”, “Lecturer”, “Lecture” and “TA Lecture”. These terms can be selected and dragged on to the canvass, where they become blogs that represent classes or individuals (for the term “Mark” in the list above). %Relationships are created by selecting a blob, choosing to create a relationship, moving to the “target” and then finishing the interaction. Relationship sub- super-class or of other types as specified by the users. To do this, SKOT takes the following approach:
- There is a canvas on which blob and line pictures can be drawn; blobs are classes and lines are relationships.
- New blobs and lines can be created on the canvas.
- Words or terms can be dragged from a word list onto the canvas, where they form new blobs.
- The diagram can be exported as OWL through the OWL API.
- SKOT projects can be saved and re-loaded.
- It is possible to load in the existential graph portion of an existing OWL ontology, extend it in SKOT and re-save it in the original file.
There’s a lot of user interface work involved in SKOT. groups of blobs can be selected and each member of the group forms a subclass relationship to a selected superclass. The layout is in the hands of the user.
James Eales used SKOT to make a toy ontology of fish, starting by just typing in a load of words about fish. This is fine, but hooking SKOT up to an automatic term recognition tool, as well as hand-typing, would be good. Once in the word list, they are ready to drag into the window where the “ontology” can be sketched.
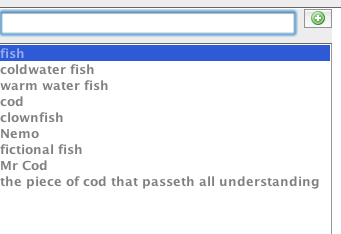
Next, James moved terms from the list on to SKOT’s sketch canvass and made a basic hierarchy of terms. Note that classes and instances are differentiated. Other types of relationship than the subclass are possible, but not used here.
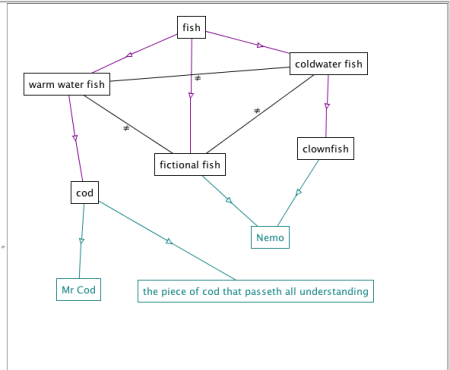
This was then saved into an OWL file, imported into Protege and shown using OWLViz.
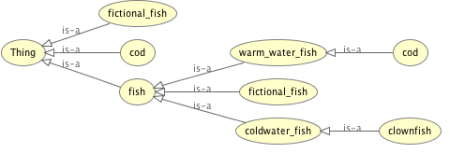
Note that the export from SKOT to OWL appears to have gone wrong – Cod is now a warm water fish, where in SKOT it was a cold water fish; I’m sure this is easily remedied.
Mark did a basic evaluation, getting some people to install and use SKOT to draft some ontologies. Two of these made ontologies — one an onmtology of guitars and one an ontology of fish. All the users were basically impressed, but also gave long lists of things to do — one user, for example, just found it difficult to work out what to do on start up; however, once he got going, all was basically OK.
SKOT is currently a stand-alone application and it really should be a Protege plugin. there’s also a lot more to do on SKOT, both little things and big things. On the list of little things are to fix various labels on the UI to make better sense. On the larger side of things, we need:
- It all connected to an automatic term recognition tool, especially with a PDF to text converter;
- We need to have regular expression searches over the term lists and editing of the list;
- We need to be able to save the list and import into the list from various sources;
- One of the main issues with SKOT is the scalability of the drawing of the blobs and lines. Some zooming would probably be useful. Montage had a facility to “fold away” portions of the sketch that wern’t currently the focus of attention. Andrew gibson had a nice design for how to deal with many of these issues, but those are not here, but some are in the Montage prototype and may see the light of day eventually. There are lots of UI tricks to be played here, but I also suspect that the utility of such a tool lies in the small scale aspect and that such things are inherently very difficult to scale.
Mark’s report on SKOT and how it was built is available.