A little while ago I wrote a blog about using an ontology or ontologies as background knowledge about a field of interest in order to learn about that domain, rather than simply annotating data or building some kind of hierarchical search interface. The idea is that an ontology captures knowledge about a field of interest; I should be able to look at that ontology and gain some kind of understanding about that domain by examining the terms used to name the class and its definition about how to recognise objects in that class (both the natural language definition and the axioms that describe that class’ objects in terms of relationships to other objects. In that blog article I conjectured that an encyclopaedia style presentation of many ontology entries could work as a way of presenting the large amount of background knowledge captured in the ontologies the community has built. My feeling is that the standard graphical presentation of blobs and lines isn’t necessarily a good way of doing this, especially when there are several ontologies at which to look. Encyclopaedia are also meant for “looking up things” and finding out about them – but we can exploit Web technologies and the structure of an ontology to get the best of both worlds. The OBO ontologies are particularly attractive for an encyclopaedia because:
- They cover a broad spectrum of biology – from sequence, through proteins, processes, functions, cells, cellular components to gross anatomy.
- Each entry in an ontology has a human readable label, synonyms, a natural language definition, all of which are”standard” parts of an encyclopaedia entry.
- The relationships between entries in the ontology can provide the “see also” links for an encyclopaedia entry.
One of my undergraduate project students for this year, Adam Nogradi, has built OBOPedia for me as an example of this kind of presentation for a group of ontologies. OBOPedia may be found via http://www.OBOPedia.org.uk. The current version of OBOPedia has nine ontologies, including the OBO Foundry ontologies plus a few more and has over 210,000 entries; the ontologies currently available are:
- The Gene Ontology.
- The Protein Ontology.
- The Chemical Entities of Biological Interest ontology.
- The Ontology of Biomedical Investigation.
- The Phenotypic Quality Ontology.
- The Zebra Fish Anatomy and Development Ontology.
- The Xenopus Anatomy and Development Ontology.
- The Human Disease Ontology.
- The Human Phenotype Ontology.
-
The Plant Ontology.
An example of an entries page for OBOPedia can be seen in the picture below:
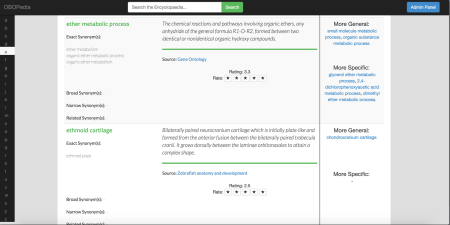
This shows Entries are arranged alphabetically. The screen here shows some entries from “E”, after some scrolling; on view are “”ether metabolic process” from GO and “ethmoid cartilage” from the Zebrafish Anatomy and Development Ontology. Each entry has the main label as the entry’s title, the various synonyms, the natural language definition and some “see also” links. The letters down the left hand side takes one to the beginning of entries starting with that letter. Entries are shown 50 at a time. One nice aspect of this style of presentation can be the serendipity of looking at entries surrounding the target entry and seeing something of interest; a typical hierarchical display automatically puts entries that are semantically related more or less in the same place – this encyclopaedia presentation doesn’t, but preserves the hierarchy via the “see also” links (though what those link to is rather hidden until arrival at the end of the link, which isn’t the case in most graphical presentations). Each entry shows the ontology whence the entry came – there are several anatomies containing the entry “lung” and knowing whence the entry comes is just a good thing. The picture also shows the (possible) exact, broader, narrower and related synonyms taken from the entries in the ontologies. At the moment OBOPedia only uses the subsumption links for “see also”s, but the aim is to expand this to other relationships in the fullness of time. I’d also like to include the ability to use DL-queries to search and filter the encyclopaedia, but time in this project has not permitted.
The picture below shows OBOPedia’s search in action.
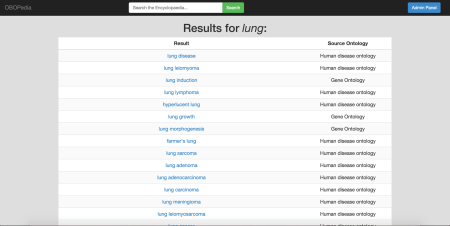
The search was for “lung” and entries were retrieved from the Gene Ontology and the Human Disease Ontology; some of the entries brought back and available for viewing were “lung disease”, “lung leiomynoma”, “lung induction”, “lung lymphoma”, “hyperlucent lung” and many others…
Along with each OBOPedia entry there is also a “rate this definition” feature. This definition rating uses a simple five point scale that allows people to rate the natural language definition (capturing comments will come at some point). The idea here is that feedback can be gathered about the natural language definitions and eventually this will form an evaluation of the definitions.
OBOPedia is an encyclopaedia of biology drawn from a subset of the OBO ontologies (there’s no reason not to include more than the nine currently on show, except for resources), exploiting their metadata, especially their natural language definitions. OBOPedia is not a text-book and it’s not a typical blob and line presentation of an ontology. It’s an encyclopaedia that presents many ontologies at once, but without the reader necessarily knowing that he or she is using an ontology. It’s an attempt to give an alternative view on the knowledge captured by the community in a range of ontologies in a way that gives easy access to that knowledge. OBOPedia may be a good thing or a bad thing. Send comments to Robert.Stevens@manchester.ac.uk or add comments to this blog post.