Simon Jupp, Robert Hoehndorf and I have realised something that we’ve been wanting to do for years and finally made the time to do (this has been written up in a JBMS paper on the Logical Gene Ontology). We’ve made annotations on mouse gene products for the Gene Ontology, Mammalian Phenotype Ontology and the Human Disease Ontologies available as a large OWL ontology that is interactively queryable on-line. (We used mouse as it is well annotated with concepts from more than one ontology.) This is the Logical Gene Ontology Annotations or GOAL. We think that it shows the utility of delivering ontologies and their queries via OWL and automated reasoning, but, perhaps more importantly, it shows that we can do this kind of thing interactively on-line; OWL tools are now sufficiently mature that this is possible.
The idea for GOAL is simple:
- We create a class of Gene product;
- For each mouse protein we create a primitive subclass of Gene product with the MGI id as URI fragment and name as label;
- Each gene product is connected to its relevant GO aspects by extracting data from the Gene Ontology Annotations;
- We also do this for the annotations from MGI for the Mammalian Phenotype Ontology and Human Disease Ontology.
- This results in a load of gene products with lots of restrictions to things we know about those proteins;
- For each of the classes in the “supporting ontologies”, we create a defined Gene product class along the lines of Class: X gene product EquivalentTo: Gene product that hasRelationshipWith some X, where “X” is the class from the supporting ontology and hasRelationshipWith is some suitable relationship;
- We classify the GOAL ontology, which is in the OWL 2 EL profile, so we can use the elk-reasoner which does the reasoning over the OWL super-fast;
- We provide a GWT based user interface to use the GOAL ontology to query these various annotations. We already have a defined class for each gene product linking it to the supporting ontology in question. Each of these defined classes recognises the various mouse gene products as appropriate. This re-builds each of the “supporting” ontologies underneath the gene product class. We can build more complex DL queries by creating intersections of two or more of these classes. So, we can ask for intra-cellular membrane bound gene products involved in abnormal cytokine secretion, are ion binding, participate in an inflammatory response with the following DL-query: x and y and z. The GOAL web page is set up to allow this. Gene product classes can be browsed to. Each gene product is “added” to the query. When complete, the “Go” button is pressed.
- The DL-query that’s been constructed is added to the ontology using the OWL API, then the ontology is re-classified by the elk-reasoner to compute all subclasses of the query, and a table of results displayed.
- For each DL-query that returns gene products, we make an addition of a defined class to the GOAL ontology. So, the query above becomes the class below as part of a “query” module that, at some point, can be added to the ontology via an import statement. So, the GOAL ontology grows lazily.
Class: `xyz gene product' EquivalentTo: x and y and z |
or for the DL query for the example above:
'immune system disease gene product' and 'abnormal cytokine secretion gene product' and 'ion binding gene product' and 'inflammatory response gene product' and 'intracellular membrane-bounded organelle gene product' |
This is a simple and straight-forward use of OWL to gain access to the rich resource of annotations of gene products with various ontologies, especially the GO. One of the tricks, just as it is with the iKUP interface to the KUPKB, is to provide some kind of reasonable user interface to the knowledge resource. I make no great claims for this user interface, but it does hide the OWL and the need to write potentially complex queries. No one should really see OWL. The OWL API gives us a good platform upon which to build applications and we have reasoners that are fast enough to do the job. The GOAL ontology has just under 150000 classes; it is in the OWL 2 EL profile, so this is really why we can do this size of ontology with the interactive, dynamic reasoning.
We’ve also been straight-forward in the ontological aspects of GOAL. We’ve said that these gene products have these functions, participate in these processes and display or are involved in these phenotypes etc. Presumably we’d have more properly said that information about these gene products has been annotated with these descriptions of functions, activities and so on. This would, I think, have added nothing to the questions being asked (apart from to make it more clumsy). I should, however, pay some thought to drawing in some of the evidence codes into this set-up, so queries can take advantages of this information. There are also things like the part_of relationships in GO that could be used with sub-property chains to say that a protein capable of a process that is part of another process is capable of the second process by implication.
The future for this work could be interesting. As well as exploring GOAL for interesting biology, we’d also like to exploit the resource to look for inconsistencies in annotation and redundancy in the annotations. This will mean adding more information to the ontology — for example, disjoints between various components. There is an increasing degree of axiomatisation in GO (and others) and it would be good to exploit this in queries. As we say in the GOAL paper, what we’ve currently done cannot detect or stop nonsense questions of gene products that cannot have a such or such location etc. Using these Gene Ontology Extensions would be a good thing.
With GOAL you can do a query like:
- Navigate down to obesity gene product (DiseaseGeneProduct > disease of metabolism gene product> acquired metabolic disease gene product > nutrition disease gene product > overnutrition gene product > obesity gene product) or simply enter obesity gene product into the search box. Press “Search” to show the results table.
- Under PhenotypeGeneProduct navigate down mammalian phenotype gene product > digestive/alimentary phenotype gene product >’abnormal digestive system physiology gene product’, and add this to the previous query. Press “Search” and view the results (See fig1).
- Select the CPE gene and select the view superclass hierarchy button. When looking at the superclasses of CPE we see it is annotated with the phenotype decreased circulating adrenocorticotropin level gene product (See fig2). Add this to the query (deleting that from step 2) to query for obesity gene product and decreased circulating adrenocorticotropin level gene product. Inspect the results.
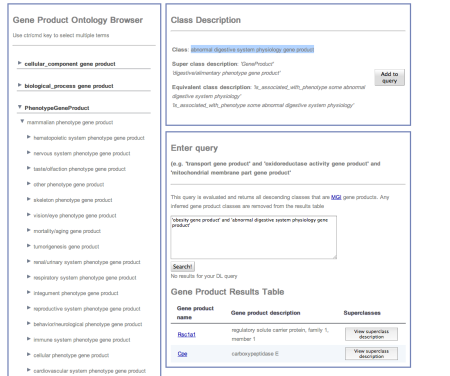
Figure 1. GOAL user interface showing query results for the DL query (obesity gene product and digestive/alimentary phenotype gene product)
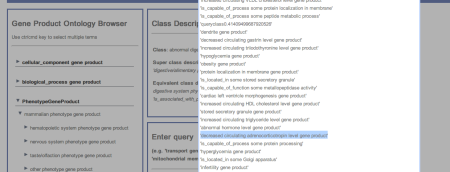
Figure 2. GOAL user interface highlighting decreased circulating adrenocorticotropin level gene product as a superclass for the DL query (obesity gene product and digestive/alimentary phenotype gene product)
The GOAL UI relies on browsing and this makes it rather clumsy. We need to add the ability to search and do things like term completion to get around the need to start from the top of each ontology and find what is wanted. Never-the-less it starts to show what can be done by adding the various ontological annotations the community has made with the OBO ontologies together to explore what could be complex biological interactions. On the technical side, it shows we can actually deliver OWL based solutions to application needs.